|
|
||
|---|---|---|
| .. | ||
| github-bitbucket | ||
| grabit-overview.png | ||
| readme.md | ||
readme.md
- What is a grabit
- Prerequisites
- Install
- Running the grabit tool
- Name of directories and files
- Configuration
- 1. SQLFlow Server
- 2. SQLScriptSource
- 3. lineageReturnFormat
- 4. databaseType
- 5. databaseServer
- hostname
- port
- username
- password
- privateKeyFile
- privateKeyFilePwd
- database
- extractedDbsSchemas
- excludedDbsSchemas
- extractedStoredProcedures
- extractedViews
- enableQueryHistory
- queryHistoryBlockOfTimeInMinutes
- queryHistorySqlType
- excludedHistoryDbsSchemas
- duplicateQueryHistory
- snowflakeDefaultRole
- metaStore
- custom ddl export sql
- 6. gitServer
- 7. SQLInSingleFile
- 8. SQLInDirectory
- 9. isUploadNeo4j
- 10. neo4jConnection
- 11. isUploadAtlas
- 12. atlasServer
- 13. donotConnectToSQLFlowServer
- 14. jobType
- Process SQL queries in a database table
- Aux features
- FAQ
What is a grabit
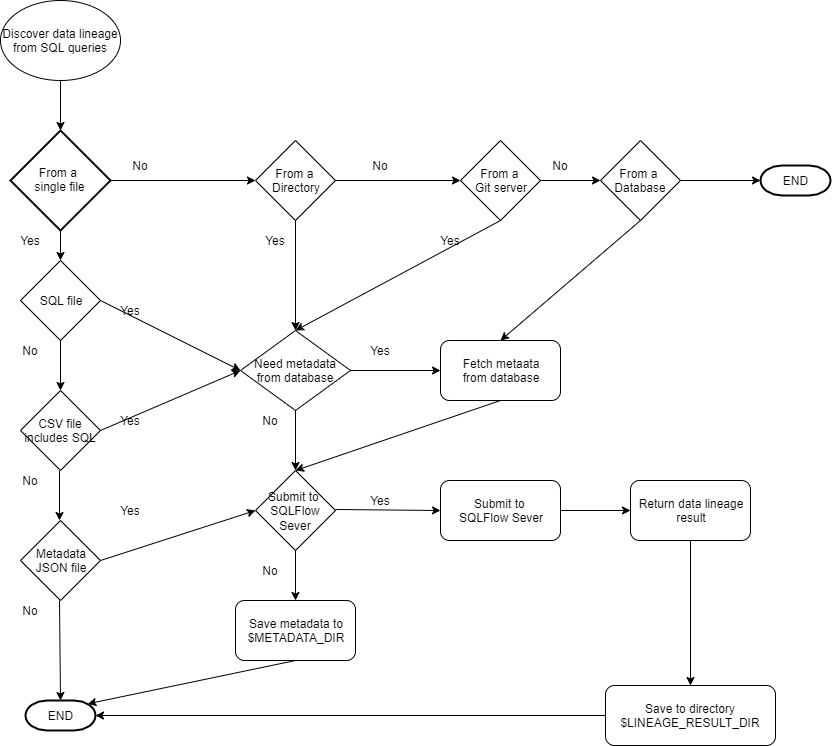
Grabit is a supporting tool for SQLFlow, which collects SQL scripts from various data sources for SQLFlow, and then uploading them to SQLFlow for data lineage analysis of these SQL scripts. The analysis results can be viewed in the browser. Meanwhile, the data lineage results will be fetched to the directory where Grabit is installed, and the JSON results can be uploaded to the Neo4j database if necessary.
Prerequisites
- Download grabit tool
- Java 8 or higher version must be installed and configured correctly.
- Grabit GUI mode only supported in Oracle Java 8 or higher version.
- Grabit Command Line mode works under both OpenJDK and Oracle JDK.
setup the PATH like this: (Please change the JAVA_HOME according to your environment)
export JAVA_HOME=/usr/lib/jvm/default-java
export PATH=$JAVA_HOME/bin:$PATH
Install
unzip grabit-x.x.x.zip
cd grabit-x.x.x
- linux & mac open permissions
chmod 777 *.sh
Running the grabit tool
The grabit tool can be running in both GUI mode and the command line mode.
GUI mode
GUI mode only runs under Oracle JDK, and only a subset of features supported in the GUI mode, The command line mode is highly recommended.
- mac & linux
./start.sh
- windows
start.bat
Command line mode
Configure file
Configure file tells the grabit tool how to collect SQL script and what's kind of data lineage result you like to achieve.
A set of pre-configed config files are located under conf-template/ directory.
You can modify it to meet your own requirement.
Running in command line
Grabit is started command-line.
- mac & linux
./start.sh -f <path_to_config_file>
note:
path_to_config_file: the full path to the config file
eg:
./start.sh -f config.txt
- windows
start.bat -f <path_to_config_file>
note:
path_to_config_file: the full path to the config file
eg:
start.bat -f config.txt
Run the grabit at a scheduled time
This guide shows you how to set up a cron job in Linux, with examples.
- use mac & linux crontab
cron ./start_job.sh -f <path_to_config_file> <lib_path>
note:
path_to_config_file: config file path
lib_path: lib directory absolute path
e.g.:
1. sudo vi /etc/crontab
2. add the following statement to the last line
1 */1 * * * ubuntu /home/ubuntu/grabit-2.4.6/start_job.sh -f /home/ubuntu/grabit-2.4.6/conf-template/oracle-config-template /home/ubuntu/grabit-2.4.6/lib
note:
0 */1 * * *: cron expression
ubuntu: The name of the system user performing the task
/home/ubuntu/grabit-2.4.6/start_job.sh: The path of the task script
-f /home/ubuntu/grabit-2.4.6/conf-template/oracle-config-template: config file path
/home/ubuntu/grabit-2.4.6/lib: lib directory absolute path
3.sudo service cron restart
Please check this document for more information about cron.
Grabit log
After execution, view the logs/graibt.log file for the detailed information.
Variable, $LOG_FILE represents the log file generated by the grabit during the execution.
$LOG_FILE = logs/graibt.log
Common Log Description
- file is not a valid file.
The file does not exist or the file address cannot be found.
- sqlScriptSource is valid, support source are database,gitserver,singleFile,directory
The sqlScriptSource parameter is incorrectly set. Data sources are only supported from databases, remote repositories,
and files and directories
- lineageReturnFormat is valid, support types are json,csv,graphml
Parameter lineageReturnFormat is incorrectly set. The data lineage result obtained can only be in JSON, CSV, and
GraphML formats
- export metadata in json successful. the resulting metadata is as follows
Exporting metadata from the specified database succeeded.
- This database is not currently supported
Parameter databaseType set error, at present only support access, bigquery, couchbase, dax, db2, greenplum, hana,
hive, impala, informix, mdx,
mssql,sqlserver,mysql,netezza,odbc,openedge,oracle,postgresql,postgres,redshift,snowflake,sybase,teradata,soql,vertica,azure
- db connect failed
The metadata fails to be exported from the specified database. If the metadata fails to be exported, check whether the
database connection information in the dataServer object is correct
- export metadata in json failed
Failed to export metadata from the specified database. Check whether the user who logs in to the database has the permission to obtain metadata
- metadata is empty
Exporting metadata from specified database is empty, please contact me for processing
- remote warehouse url cannot be empty
The URL in the gitServer parameter cannot be empty
- remote warehouse pull failed
Failed to connect to the remote warehouse. Check whether the remote warehouse connection information is correct
- connection failed,repourl is the ssh URL
The remote repository address is incorrect. Please check whether it is a Git address
- remote warehouse file to zip successful. path is:xx
Pull to a local storage address from a remote repository
- get token from sqlflow failed
Failed to connect to SQLFlow. Check whether connection parameters of sqlflowServer are correct
- submit job to sqlflow failed, please input https with url
Failed to submit the SQLFlow task. Check whether the URL and port of the sqlflowServer are correct
- submit job to sqlflow failed
Failed to submit the SQLFLOW task. Check whether the sqlFLOW background service is started properly
- get job to status failed
After a job is submitted to SQLFLOW, SQLFlow fails to execute the job
- export json result failed
Description Failed to export Data Lineage in JSON format from SQLflow
- export csv result failed
Description Failed to export Data Lineage in csv format from SQLflow
- export diagram result failed
Description Failed to export Data Lineage in diagram format from SQLflow
- submit job to sqlflow successful
The job is successfully submitted to SQLFlow, and the basic information about the submitted job is displayed
- [database: 0 table: 0 view: 0 procedure: 0 column: 0 synonym: 0]
Statistics the amount of metadata exported from the specified database
- the time taken to export : 0ms
Time, in milliseconds, to export metadata from the specified database
- download success, path: xxx
Local storage address of Data Lineage returned after successfully submitting a job to SQLFlow
- job id is : xxxx
job id from sqlflow , log in to the SQLFlow website to view the newly analyzed results. In the Job List, you can view
the analysis results of the currently submitted tasks.
- The number of relationships in this task is too large to export this file, please check blood relationships on SQLFlow platform.
When the task uploaded to SQLFlow is too large and the number of rolls parsed by SQLFlow is too large, Grabit cannot obtain CSV files from it and needs to check the blood relationship of this task on SQLFlow.
Name of directories and files
- The name of submitted job is
grabit_%yyyyMMddHHmmss%, Variable$JOB_NAMErepresents the job name.
$JOB_NAME = grabit_%yyyyMMddHHmmss%
- After export metadata from the database, the metadata data is saved under the
data/job_%jobname%/metadatadirectory. Variable$METADATA_DIRrepresents the directory when the metadata exported from a database is saved.
$METADATA_DIR = data/job_$JOB_NAME/metadata
Metadata file name: metadata.json
- Once the job is done, the data lineage result generated by the SQLFlow is saved under the
data/job_%jobname%/resultdirectory. Variable:$LINEAGE_RESULT_DIRrepresents the directory where the lineage result of a job is saved.
$LINEAGE_RESULT_DIR = data/job_$JOB_NAME/result
Lineage result file name: data-lineage-result.json
lineageReturnOutputFile
The value of $LINEAGE_RESULT_DIR will be overwrited by the value of lineageReturnOutputFile which
specify the data lineage result file directly.
"lineageReturnOutputFile":"/user/data.csv"
PLEASE NOTE THAT ALL VARIABLE NAMES THAT START WITH $ SIGN IN THIS SECTION IS USED FOR COMMUNICATION ONLY, THEY DO NOT BEEN USED IN THE CONFIGURE FILE!
Configuration
Modify the configure file to set all parameters correctly according to your environment.
1. SQLFlow Server
This is the SQLFlow server that the grabit sends the SQL script.
server
Usually, it is the IP address of the SQLFlow on-premise version
installed on your owner servers such as 127.0.0.1 or http://127.0.0.1
You may set the value to https://api.gudusoft.com if you like to send your SQL script
to the SQLFlow Cloud Server to get the data lineage result.
serverPort
The default value is 8081 if you connect to your SQLFlow on-premise server.
However, if you setup the nginx reverse proxy in the nginx configuration file like this:
location /api/ {
proxy_pass http://127.0.0.1:8081/;
proxy_connect_timeout 600s ;
proxy_read_timeout 600s;
proxy_send_timeout 600s;
proxy_set_header Host $host;
proxy_set_header X-Real-IP $remote_addr;
proxy_set_header X-Forwarded-For $proxy_add_x_forwarded_for;
proxy_set_header User-Agent $http_user_agent;
}
Then, keep the value of serverPort empty and set server to the value like this: http://127.0.0.1/api.
Please keep this value empty if you connect to the SQLFlow Cloud Server by specifying the
https://api.gudusoft.comin theserver
userId userSecret
This is the user id that is used to connect to the SQLFlow server. Always set this value to gudu|0123456789 and
keep userSecret empty if you use the SQLFlow on-premise version.
If you want to connect to the SQLFlow Cloud Server, you may request a 30 days premium account to get the necessary userId and secret code.
Example configuration for on-premise version:
"SQLFlowServer":{
"server": "127.0.0.1",
"serverPort": "8081",
"userId": "gudu|0123456789",
"userSecret": ""
}
Example configuration for Cloud version:
"SQLFlowServer":{
"server": "https://api.gudusoft.com",
"serverPort": "",
"userId": "your own user id here",
"userSecret": "your own secret key here"
}
2. SQLScriptSource
You may collect SQL scripts from various sources such as database, Github repo, file system. This parameter tells grabit where the SQL scripts come from.
Available values for this parameter:
- database
- gitserver
- singleFile
- directory
This configuration means the SQL script is collected from a database.
"SQLScriptSource":"database"
2.1. enableGetMetadataInJSONFromDatabase
If the source of the SQL scripts is not the database, we may specify a database by setting databaseServer parameter to
fetch metadata from the database instance to help SQLFlow get a more accurate result during the analysis.
If enableGetMetadataInJSONFromDatabase=1, You must set all necessary information in databaseServer.
Of course, you can enableGetMetadataInJSONFromDatabase=0. This means all SQL scripts will be analyzed offline without
a connection to the database. SQLFlow still works quite well to get the data lineage result by taking advantage of its
powerful SQL analysis capability.
Sample configuration of enable fetching metadata in json from the database:
"enableGetMetadataInJSONFromDatabase":1
3. lineageReturnFormat
When you submit SQL script to the SQLFlow server, A job is created on the SQLFlow server and you can always see the graphic data lineage result in the frontend of the SQLFlow by using the browser,
Even better, grabit will fetch the data lineage back to the directory where the grabit is running. Those data lineage
results are stored in the data/job_%jobname/result/ directory.
This parameter specifies which kind of format is used to save the data lineage result.
Available values for this parameter:
- json, data lineage result in JSON.
- csv, data lineage result in CSV format.
- graphml, data in graphml format that can be viewed by yEd.
This sample configuration means the output format is json.
"lineageReturnFormat":"json"
4. databaseType
This parameter specifies the database dialect of the SQL scripts that the SQLFlow has analyzed.
access,bigquery,couchbase,dax,db2,greenplum,hana,hive,impala,informix,mdx,mssql,
sqlserver,mysql,netezza,odbc,openedge,oracle,postgresql,postgres,redshift,snowflake,
sybase,teradata,soql,vertica,azure
This sample configuration means the SQL dialect is SQL Server database.
"databaseType":"sqlserver"
5. databaseServer
This option is used when
-
- Need to discover data lineage of a specific database:
SQLScriptSource=database
- Need to discover data lineage of a specific database:
-
- Need to discover lineage from SQL in a text file, but need metadata from a database in order to get a more accurate result.
-
- Need to discover the Hive metaStore that stored in a database.
hostname
The IP of the database server that the grabit connects.
port
The port number of the database server that the grabit connect.
username
The database user used to login to the database.
password
The password of the database user.
note: the passwords can be encrypted using tools [Encrypted password](#Encrypted password), using encrypted passwords more secure.
privateKeyFile
Use a private key to connect, Only supports the snowflake.
privateKeyFilePwd
Generate the password for the private key, Only supports the snowflake.
database
The name of the database instance to which it is connected.
For azure,greenplum,netezza,oracle,postgresql,redshift,teradata databases, it represents the database name and is required, For other databases, it is optional.
note: If this parameter is specified and the database to which it is connected is Azure, Greenplum, PostgreSQL, or Redshift, then only metadata under that library is extracted.
extractedDbsSchemas
List of databases and schemas to extract, separated by commas, which are to be provided in the format database/schema;
Or blank to extract all databases.
database1/schema1,database2/schema2,database3 or database1.schema1,database2.schema2,database3
When parameter database is filled in, this parameter is considered a schema. And support wildcard characters such
as database1/*,*/schema,*/*.
When the connected databases are Oracle and Teradata, this parameter is set the schemas, for example:
extractedDbsSchemas: "HR,SH"
When the connected databases are Mysql , Sqlserver, Postgresql, Snowflake, Greenplum, Redshift, Netezza
, Azure, this parameter is set database/schema, for example:
extractedDbsSchemas: "MY/ADMIN"
excludedDbsSchemas
This parameters works under the resultset filtered by extractedDbsSchemas. List of databases and schemas to exclude
from extraction, separated by commas
database1/schema1,database2 or database1.schema1,database2
When parameter database is filled in, this parameter is considered a schema. And support wildcard characters such
as database1/*,*/schema,*/*.
When the connected databases are Oracle and Teradata, this parameter is set the schemas, for example:
excludedDbsSchemas: "HR"
When the connected databases are Mysql , Sqlserver, Postgresql, Snowflake, Greenplum, Redshift, Netezza
, Azure, this parameter is set database/schema, for example:
excludedDbsSchemas: "MY/*"
extractedStoredProcedures
A list of stored procedures under the specified database and schema to extract, separated by commas, which are to be
provided in the format database.schema.procedureName or schema.procedureName; Or blank to extract all databases, support
expression.
database1.schema1.procedureName1,database2.schema2.procedureName2,database3.schema3,database4
or database1/schema1/procedureName1,database2/schema2
for example:
extractedStoredProcedures: "database.scott.vEmp*"
or
extractedStoredProcedures: "database.scott"
extractedViews
A list of stored views under the specified database and schema to extract, separated by commas, which are to be provided
in the format database.schema.viewName or schema.viewName. Or blank to extract all databases, support expression.
database1.schema1.procedureName1,database2.schema2.procedureName2,database3.schema3,database4
or database1/schema1/procedureName1,database2/schema2
for example:
extractedViews: "database.scott.vEmp*"
or
extractedViews: "database.scott"
enableQueryHistory
Fetch SQL queries from the query history if set to true default is false.
queryHistoryBlockOfTimeInMinutes
When enableQueryHistory:true, the interval at which the SQL query was extracted in the query History,default is 30
minutes.
queryHistorySqlType
When enableQueryHistory:true, the DML type of SQL is extracted from the query History. When empty, all types are
extracted, and when multiple types are specified, a comma separates them, such as SELECT,UPDATE,MERGE. Currently only
the snowflake database supports this parameter,support types are SHOW,SELECT,INSERT,UPDATE,DELETE,MERGE,CREATE TABLE,
CREATE VIEW, CREATE PROCEDURE, CREATE FUNCTION.
for example:
queryHistorySqlType: "SELECT,DELETE"
excludedHistoryDbsSchemas
This parameters works under the resultset filtered by excludedHistoryDbsSchemas. List of databases and schemas to
exclude from extraction, separated by commas database1.schema1,database2. for example:
excludedHistoryDbsSchemas: "DB1.SCHEMA1,DB2.SCHEMA2"
duplicateQueryHistory
When duplicateQueryHistory:1, duplicate History Queries are filtered, not filtered by default 0.
snowflakeDefaultRole
This value represents the role of the snowflake database.
note: You must define a role that has access to the SNOWFLAKE database,And assign WAREHOUSE permission to this role.
Assign permissions to a role, for example:
#create
role
use role accountadmin;
grant imported
privileges on database snowflake to role sysadmin;
grant imported
privileges on database snowflake to role customrole1;
use
role customrole1;
select *
from snowflake.account_usage.databases;
#To
do this, the Role gives the WAREHOUSE permission
select current_warehouse()
use role sysadmin GRANT ALL PRIVILEGES
ON WAREHOUSE %current_warehouse% TO ROLE customrole1;
Please refer to:Grant Privileges
metaStore
ref #: I44EE8
This option means the metadata is fetched from a Hive or SparkSQL metaStore. The metaStore uses the RDBMS such as MySQL to save the metadata.
Only Hive metaStore is supported in current version.
when this option is set to hive, Grabit extract metadata from the metaStore,
but not from the common metadata of the database.
Sample configuration of a SQL Server database:
"hostname":"127.0.0.1",
"port": "1433",
"username": "sa",
"password": "PASSWORD",
"database": "",
"extractedDbsSchemas":"AdventureWorksDW2019/dbo",
"excludedDbsSchemas": "",
"extractedStoredProcedures": "AdventureWorksDW2019.dbo.f_qry*",
"extractedViews": "",
"enableQueryHistory": false,
"queryHistoryBlockOfTimeInMinutes":30,
"snowflakeDefaultRole": "",
"queryHistorySqlType": "",
"metaStore": "hive"
custom ddl export sql
conf.zip file contains all ddl export sql, you can edit the sql file in the conf.zip, keep the same of return
fields, put the modified sql file at: conf/%database_name%/%query_type%.sql
for example, when you edit the conf/mssql/query.sql, please copy it to conf/mssql/query.sql, the grabit will load your modified sql file as ddl export sql.
Please check conf.zip download.
6. gitServer
When SQLScriptSource=gitserver, grabit will fetch SQL files from a specified github or bitbucket repo, the SQL script
files are stored in data/job_%jobname%/metadata/ before submitting to the SQLFlow server.
Both sshkey and account password authentication methods are supported.
url
Pull the repository address of the SQL script from GitHub or BitBucket.
note: If sshkey authentication is used, you must enter the SSH address.
username
Pull the user name to which the SQL script is connected from GitHub or BitBucket.
password
Pull the personal token to which the SQL script is connected from GitHub or BitBucket.
sshKeyPath
The full path to the SSH private key file.
Sample configuration of the GitHub or BitBucket public repository servers :
"url":"your public repository address here",
"username": "",
"password": "",
"sshKeyPath": ""
Sample configuration of the GitHub or BitBucket private repository servers:
"url":"your private library address here",
"username": "your private repository username here",
"password": "your private repository personal token here",
"sshKeyPath": ""
or
"url":"your private repository ssh address here",
"username": "",
"password": "",
"sshKeyPath": "your private repository ssh key address here"
7. SQLInSingleFile
When SQLScriptSource=singleFile, this is a single SQL file needs to be analyzed.
- filePath
The name of the SQL file with full path.
- csvFormat
Format of a CSV file. used to represent the CSV in the Catalog, Schema, ObjectType, ObjectName, ObjectCode, Notes
each column is the number of columns in the CSV file, does not exist it is 0, The default is 123456.
- objectCodeEncloseChar
Specifies that the string contains SQL Code content.
- objectCodeEscapeChar
ObjectCodeEncloseChar specifies the string escape.
- redshiftLog
The default value of redshiftLog is 0, indicating whether the uploaded file is a common SQL script file. For example, if the redshift log file needs to be parsed by SQLFlow, set the value to 1.
- isDumpDDL
The default value of isDumpDDL is 0, this value is set to 1 when the specified file is a DDL file from the database dump.
- databaseName
The default value of databaseName is left blank. When the specified file is a DDL file from a database dump, this value is set to the name of the database from which the DDL was retrieved.
8. SQLInDirectory
When SQLScriptSource=directory, SQL files under this directory including sub-directory will be analyzed.
- directoryPath
The directory includes the SQL files.
- redshiftLog
The default value of redshiftLog is 0, indicating whether the uploaded file is a common SQL script file. For example, if the redshift log file needs to be parsed by SQLFlow, set the value to 1.
9. isUploadNeo4j
Upload the data lineage result to a Neo4j database for further processing. Available values for this parameter is 1 or 0, enable this function if the value is 1, disable it if the value is 0, the default value is 0.
Sample configuration of a Whether to upload neo4j:
"isUploadNeo4j":1
10. neo4jConnection
If IsuploadNeo4j is set to '1', this parameter specifies the details of the neo4j server.
- url
The IP of the neo4j server that connects to.
- username
The user name of the neo4j server that connect to.
- password
The password of the neo4j server that connect to.
Sample configuration of a local directory path:
"url":"127.0.0.1:7687",
"username": "your server username here",
"password": "your server password here"
11. isUploadAtlas
Upload the metadata to a Atlas server for further processing. Available values for this parameter is 1 or 0, enable this function if the value is 1, disable it if the value is 0, the default value is 0.
Sample configuration of a Whether to upload neo4j:
"isUploadAtlas":1
12. atlasServer
If isUploadAtlas is set to '1', this parameter specifies the details of the atlas server.
- ip
The IP of the atlas server that connects to.
- port
The PORT of the atlas server that connects to.
- username
The user name of the atlas server that connect to.
- password
The password of the atlas server that connect to.
Sample configuration of a local directory path:
"ip":"127.0.0.1",
"port": "21000",
"username": "your server username here",
"password": "your server password here"
13. donotConnectToSQLFlowServer
If donotConnectToSQLFlowServer is set to 1, the metadata file is not uploaded to SQLFlow. the default is 0
14. jobType
If jobType is set to "regular", tasks are submitted to SQLFlow incrementally and in batches according to reualr mode. the default is "simple"
eg configuration file:
{
"databaseServer": {
"hostname": "127.0.0.1",
"port": "1433",
"username": "sa",
"password": "PASSWORD",
"privateKeyFile": "",
"privateKeyFilePwd": "",
"database": "",
"extractedDbsSchemas": "AdventureWorksDW2019/dbo",
"excludedDbsSchemas": "",
"extractedStoredProcedures": "",
"extractedViews": "",
"enableQueryHistory": false,
"excludedHistoryDbsSchemas":"" ,
"duplicateQueryHistory":0 ,
"queryHistoryBlockOfTimeInMinutes": 30,
"snowflakeDefaultRole": "",
"queryHistorySqlType": "",
"metaStore": ""
},
"gitServer": {
"url": "https://github.com/sqlparser/snowflake-data-lineage",
"username": "",
"password": "",
"sshkeyPath": ""
},
"SQLInSingleFile": {
"filePath": "",
"csvFormat": "",
"objectCodeEncloseChar": "",
"objectCodeEscapeChar": "",
"dumpDDL": 0,
"databaseName": ""
},
"SQLInDirectory": {
"directoryPath": ""
},
"SQLFlowServer": {
"server": "http:127.0.0.1",
"serverPort": "8081",
"userId": "gudu|0123456789",
"userSecret": ""
},
"neo4jConnection": {
"url": "",
"username": "",
"password": ""
},
"atlasServer": {
"ip": "127.0.0.1",
"port": "21000",
"username": "",
"password": ""
},
"isUploadAtlas": 0,
"SQLScriptSource": "database",
"lineageReturnFormat": "json",
"lineageReturnOutputFile": "",
"databaseType": "snowflake",
"isUploadNeo4j": 0,
"donotConnectToSQLFlowServer": 0,
"enableGetMetadataInJSONFromDatabase": 0,
"jobType": "simple"
}
Process SQL queries in a database table
Ref #: I43QRV
This feature will extract SQL queries saved in a database table, metadata of the same database will also be extracted into the same JSON file.
Option also need to be set is databaseServer.
sqlsourceTableName
Name of the table where SQL queries are saved.
table name: query_table
| query_name | query_source |
|---|---|
| query1 | create view v1 as select f1 from t1 |
| query2 | create view v2 as select f2 from t2 |
| query3 | create view v3 as select f3 from t3 |
If you save SQL queries in a specific table, one SQL query per row.
Let's say: The column query_table.query_source stores the source code of the query.
We can use this query to fetch all SQL queries in this table:
select query_name as queryName, query_source as querySource
from query_table
By setting the value of sqlsourceTableName and sqlsourceColumnQuerySource,sqlsourceColumnQueryName
grabit can fetch all SQL queries in this table and send it to the SQLFlow to analzye the lineage.
In this example,
"sqlsourceTableName":"query_table"
"sqlsourceColumnQuerySource":"query_source"
"sqlsourceColumnQueryName":"query_name"
Please leave sqlsourceTableName empty if you don't fetch SQL queries from a specific table.
sqlsourceColumnQuerySource
In the above sample:
"sqlsourceColumnQuerySource":"query_source"
sqlsourceColumnQueryName
"sqlsourceColumnQueryName":"query_name"
This parameter is optional, you don't need to speicify a query name column if it doesn't exist in the table.
Aux features
Extract queries that surrounded by the single quote from any files
Read text files from a specified directory, extract text that surrounded by the single quote character from each file in the directory and put the text in a separate new file if the text represents a valid SQL statement.
reference #: I48FCX
- mac & linux
./start.sh -e --generic -t dbvendor dir_to_txt_file
eg:
./start.sh -e --generic -t oracle /root/oracledir
- windows
start.bat -e --generic -t dbvendor dir_to_txt_file
eg:
start.bat -e --generic -t oracle /root/oracledir
Extract queries in metadata jsosn file to a new sql file
After exporting metadata from a database, the metadata is saved in a JSON file. Queries such as create view are included in this JSON file.
By using this option, grabit will extracts all queries in the JSON file, and put each query in a separate text file.
Ref #: I3DTWP
- mac & linux
./start.sh -e path_to_json_file [--targetDir target_dir]
note:
path_to_config_file: the full path to the metedata json file
target_dir: the path to the generated SQL file, optional
eg:
./start.sh -e test.json --targetDir /root/sqlfiles
- windows
start.bat -e path_to_json_file [--targetDir target_dir]
note:
path_to_config_file: the full path to the metedata json file
target_dir: the path to the generated SQL file, optional
eg:
start.bat -e test.json --targetDir /root/sqlfiles
Export metadata in csv to sql files
Extract SQL queries in a CSV file and save each SQL query in a separate text file.
Detailed descrition of the CSV file format, please check this document
Ref #: I45ANU
- mac & linux
./start.sh -e --csv path_to_csv_file [--csvFormat 123456] [--objectCodeEncloseChar char] [--objectCodeEscapeChar char] [--targetDir target_dir]
note:
path_to_config_file: the full path to the metedata csv file
target_dir: the path to the generated SQL file, optional
eg:
./start.sh -e --csv test.csv --csvFormat 123456 --objectCodeEscapeChar " --objectCodeEncloseChar " --targetDir /root/sqlfiles
- windows
start.bat -e --csv path_to_csv_file [--csvFormat 123456] [--objectCodeEncloseChar char] [--objectCodeEscapeChar char] [--targetDir target_dir]
note:
path_to_config_file: the full path to the metedata csv file
target_dir: the path to the generated SQL file, optional
eg:
start.bat -e --csv test.csv --csvFormat 123456 --objectCodeEscapeChar " --objectCodeEncloseChar " --targetDir /root/sqlfiles
Encrypted password
Encrypt the database connection password.
- mac & linux
./start.sh --encrypt password
note:
password: the database connection password
eg:
./start.sh --encrypt 123456
- windows
./start.bat --encrypt password
note:
password: the database connection password
eg:
./start.bat --encrypt 123456
FAQ
- Q: Is it possible to upload multiple scripts and view the lineage across these scripts in a combined way?
A: Yes, you can add multiple scripts to a single zip file and then upload this zip file to SQLFlow by creating a job